Unlike JDBC, ODBC is language-independent. ODBC is an open interface that can be used to communicate with databases. All you should do is create an ODBC connection to EXASOL database in your system. For windows, follow these steps to add ODBC creation. Here is an example of using EXASOL data for predictive analysis in R tool.
Steps to Create ODBC Connection
- Click on the Start Menu
- Select Control Panel
- Select Administrative Tools and double-click the ODBC Data Sources (either 32-bit or 64-bit) icon
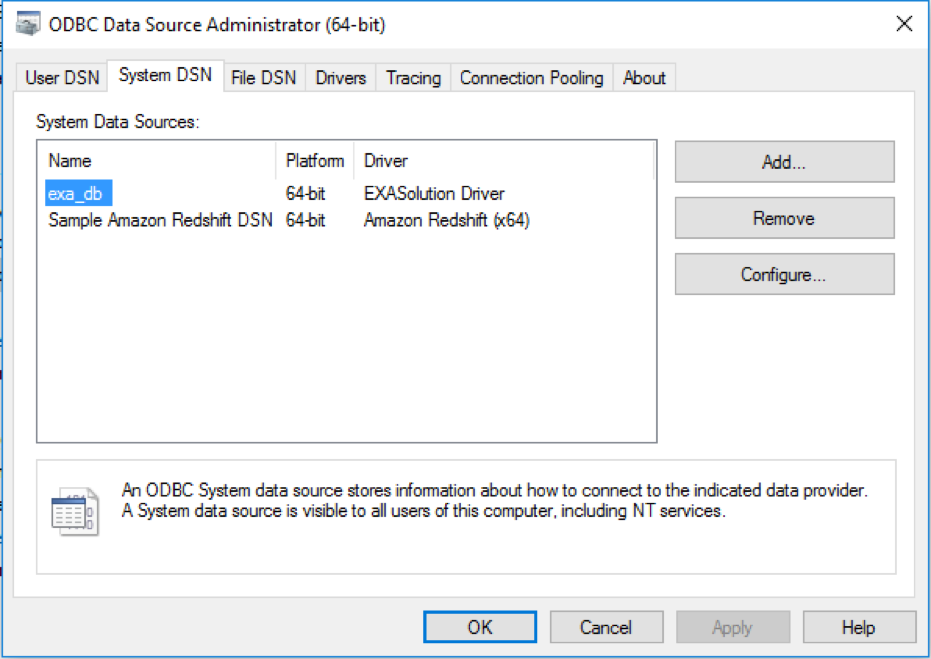
- Click on the System DSN tab
- Click the Add button
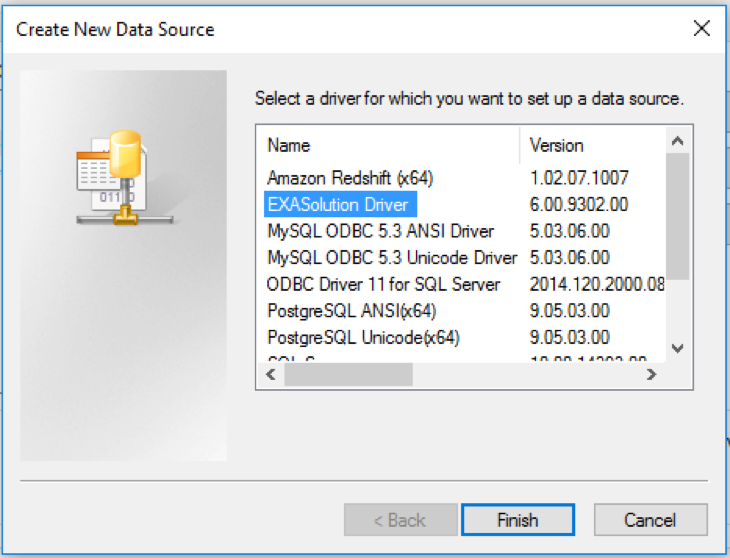
- Select EXASolution Driver from the driver’s list and click the Finish button
How To Import Data Into EXASOL
In this example, I am using two datasets: train and test. Train dataset is used to build a predictive model and test dataset is used to validate or test the model built. Do not panic if you do not have datasets handy in EXASOL. All you should do is use import statements to load data from your local machine. An import statement in EXASOL can get you gigabytes of data in a few minutes.
I used the import statement for the first time and it was so fast. It seemed like the dataset was already there in EXASOL and I didn’t need to do any importing. Remember to create tables in EXASOL as per the dataset before importing the data.
I already have the train.csv and test.csv file in C: drive and I am simply giving the path to import. Since it is a CSV file, column separator is “comma.”
IMPORT INTO retail.TRAIN FROM LOCAL CSV FILE ‘C:train.csv’ ENCODING = ‘UTF-8’ ROW SEPARATOR = ‘CRLF’ COLUMN SEPARATOR = ‘,’ COLUMN DELIMITER = ‘”’ SKIP = 1 REJECT LIMIT 0; IMPORT INTO retail.TEST FROM LOCAL CSV FILE ‘C:test.csv’ ENCODING = ‘UTF-8’ ROW SEPARATOR = ‘CRLF’ COLUMN SEPARATOR = ‘,’ COLUMN DELIMITER = ‘”’ SKIP = 1 REJECT LIMIT 0;
EXASOL Data To Build Predictive Models In R
First, you need to install EXASOL package in R. The package can be found on GitHub. This example demonstrates the use of the random forest technique to predict image id in test data given the pixel input.
# packages
install.packages("RODBC")
library(RODBC)
install.packages("randomForest")
library(randomForest)
install.packages("devtools")
devtools::install_github("exasol/r-exasol")
Connect to EXASOL using the ODBC connection that was made in the first step and read the data from tables in EXASOL.
# connect
C <- odbcConnect("exa_db")
# read data
train <- exa.readData(C, "SELECT * FROM retail.TRAIN")
test <- exa.readData(C, "SELECT * FROM retail.TEST")
Remove image id from the test data since we are using this data to predict values of image id using the model. Also, extract the labels from the train data. “Labels” in the train data is simply the target variable.
# remove imageid from test test <- test[order(test[1]),] imageid <- as.factor(test[,1]) test <- test[,-1] # extract labels from train labels <- as.factor(train[,1]) train <- train[,-1]
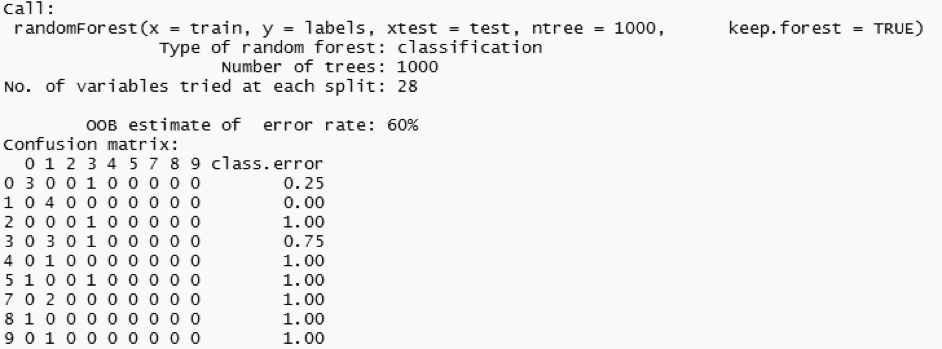
I am using the random forest technique to build 500 trees. Random forests correct overfitting problems created by decision trees, because random forests are an ensemble method of classification that operates by constructing many decision trees.
# random forest algorithm model <- randomForest(train, labels, xtest=test, ntree=500, keep.forest=TRUE) predictions <- cbind(imageid ,levels(labels)[rf$test$predicted]) # print prediction predictions # print forest model
You can also save the model built locally and reuse it with a different dataset.
# save the model locally
save(model,file="model")
# reload and reuse
load("model")
predict(model,test)
The model turned out to have a misclassification rate of 60%. We can see that the predicted values for image id four to nine are wrong. The model could definitely be trained in conjunction with larger datasets. This is where EXASOL comes in handy since we can load and access huge datasets within mere seconds.